Ribonucleic acid (RNA) is an elemental component of biological cells. While deoxyribonucleic acid (DNA) is the storage medium of genetic information, RNA has a much more complex biochemical functionality. This includes the transfer of information in the form of mRNA, RNA-mediated catalytic activity in ribosomes to the storage of genetic information in viruses. Chemically, RNA consists of a sequence of organic nucleobase molecules that are held together by a so-called backbone of phosphate and sugar groups. Such a molecular strand may be present singly or in the form of a double helix. Both macromolecular forms are embedded in a water envelope; the oxygen atoms of the phosphate and sugar groups are excellent contact points for water molecules. The structure of the water envelope has fluctuations in a time range of fractions of a picosecond (1 ps = 10-12 s = 1 millionth of a millionth of a second). The interaction between RNA and water and their role in the formation of three-dimensional RNA structures are only partially understood and difficult to access experimentally.
Flexibility and order - the interaction between ribonucleic acid and water
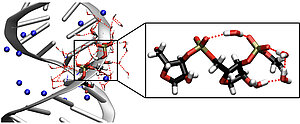
Fig. 1: Left: structure of the RNA double helix, blue spheres represent sodium counter ions. Right: enlargement of the RNA backbone consisting of phosphate and sugar groups, bridging water molecules are shown schematically. The vibrations of the RNA backbone serve as sensitive probes to track the influence of immediately adjacent water molecules on the structure and dynamics of RNA in real time.
Researchers at the Max Born Institute have now followed the interaction between RNA and the surrounding water envelope in real time using a new experimental method. Vibrations of the RNA backbone serve as sensitive probes for the influence of the immediately adjacent water molecules on the structure and dynamics of the RNA. With the so-called two-dimensional vibration spectroscopy, the temporal development of vibrational excitations can be recorded and molecular interactions within the RNA as well as between RNA and water can be determined. It turns out that water molecules on the RNA surface perform ultra-fast tilting movements in fractions of a picosecond, but maintain their local spatial arrangement for a period of more than 10 picoseconds. This behavior deviates significantly from the dynamics of pure water and is strongly influenced by the spatial boundary conditions at the RNA surface. Individual water molecules combine neighboring phosphate groups and form a partially ordered structure, which is mediated by coupling to the sugar moieties.
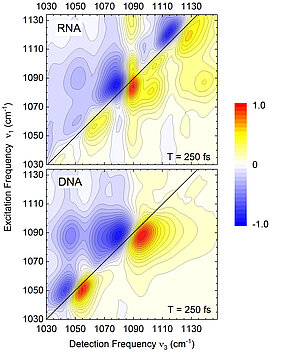
Fig. 2: Two-dimensional vibrational spectra of RNA (top) and DNA (bottom) in the frequency range of the sugar-phosphate vibrations of the backbone. The RNA spectrum has additional bands (contours) along the frequency diagonal ν1 = ν3 and a more complex distribution of extra-diagonal contributions. In addition to the frequency positions, the line shapes of the individual bands (contours) give information about details of the interaction with neighboring water molecules.
The moving water molecules generate an electric force, with which the water fluctuations are transferred to vibrations of the RNA. The vibrations of the RNA backbone show a different dynamic behavior, which is determined by the local water environment and reflects their heterogeneity. RNA oscillations, in turn, couple to each other, exchanging energy with each other and with the water envelope. The associated ultrafast redistribution of excess energy prevents local overheating of the macromolecular structure. This complex scenario has been analyzed by detailed theoretical calculations and simulations, which include: The vibrational movements of the RNA backbone were completely and quantitatively identified for the first time. Comparative experiments on DNA reveal similarities as well as characteristic differences in the behavior of these two elementary biomolecules, with RNA characterized by a more structured arrangement of the surrounding water envelope. The results of the study demonstrate the diverse potential of non-invasive time-resolved vibrational spectroscopy to decipher the interplay of structure and dynamics on molecular length and time scales in complex biomolecular systems.
Search publications of MBI
Publications since 2025